

BiOFiLM is a computer program to design deep-biofilm packed beds that runs under Windows 95. The program is used to calculate the bioreactor volume required to achieve the desired removal of the substrate of interest. In this context, the substrate of interest may be either the electron donor, as in the case of carbonaceous oxidation (substrate = BOD), or the electron acceptor, as in the case of denitrification (substrate = NO3-N).
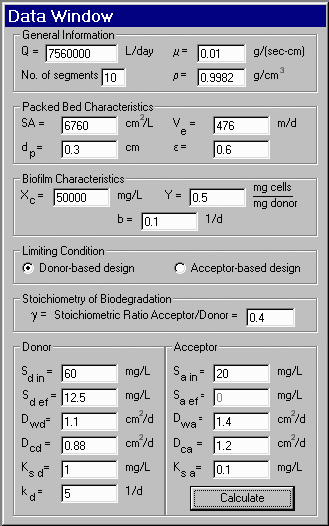
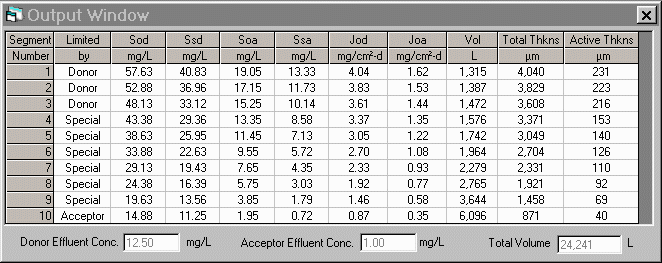
The bioreactor is calculated based
on the physical and biological parameters provided by the user. A sample
of the input and output windows of the program is presented below.
 |
 |
Besides calculating the size of
a bioreactor, the program can produce graphs that can help explore and
understand the behavior of deep-biofilm reactors. Below is a sample of
the graphs that BiOFiLM can produce.
|
|
|
|
|
|
BiOFiLM can be downloaded online using the links below. If you have Visual Basic 5.0 or higher installed in your computer, you can use link 1; otherwise you must use link 2 since it includes the runtime libraries required by BiOFiLM.
BiOFiLM comes in compressed zip
files, so you will need a decompression utility such as PKUNZIP or WINZIP
to expand the installation files.
After extracting the files, BiOFiLM is ready to use, and you can start it from the Windows Explorer, or you might want to create a shortcut for it on your desktop.
For more information about BiOFiLM
contact Dr. Gerald E. Speitel Jr. (speitel@mail.utexas.edu).
Back
to Homepage
Back to Civil
Engineering Homepage
Back to UT Homepage